“Background: Recent approved medicines whose active principles are Δ9Tetrahidrocannabinol (Δ9-THC) and/or cannabidiol (CBD) open novel perspectives for other phytocannabinoids also present in Cannabis sativa L. varieties. Furthermore, solid data on the potential benefits of acidic and varinic phytocannabinoids in a variety of diseases are already available. Mode of action of cannabigerol (CBG), cannabidiolic acid (CBDA), cannabigerolic acid (CBGA), cannabidivarin (CBDV) and cannabigerivarin (CBGV) is, to the very least, partial.
“Background: Recent approved medicines whose active principles are Δ9Tetrahidrocannabinol (Δ9-THC) and/or cannabidiol (CBD) open novel perspectives for other phytocannabinoids also present in Cannabis sativa L. varieties. Furthermore, solid data on the potential benefits of acidic and varinic phytocannabinoids in a variety of diseases are already available. Mode of action of cannabigerol (CBG), cannabidiolic acid (CBDA), cannabigerolic acid (CBGA), cannabidivarin (CBDV) and cannabigerivarin (CBGV) is, to the very least, partial.
Hypothesis/purpose: Cannabinoid CB1 or CB2 receptors, which belong to the G-protein-coupled receptor (GPCR) family, are important mediators of the action of those cannabinoids. Pure CBG, CBDA, CBGA, CBDV and CBGV from Cannabis sativa L. are differentially acting on CB1 or CB2 cannabinoid receptors.
Study design: Determination of the affinity of phytocannabinoids for cannabinoid receptors and functional assessment of effects promoted by these compounds when interacting with cannabinoid receptors.
Methods: A heterologous system expressing the human versions of CB1 and/or CB2 receptors was used. Binding to membranes was measured using radioligands and binding to living cells using a homogenous time resolved fluorescence resonance energy transfer (HTRF) assay. Four different functional outputs were assayed: determination of cAMP levels and of extracellular-signal-related-kinase phosphorylation, label-free dynamic mass redistribution (DMR) and ß-arrestin recruitment.
Results: Affinity of cannabinoids depend on the ligand of reference and may be different in membranes and in living cells. All tested phytocannabinoids have agonist-like behavior but behaved as inverse-agonists in the presence of selective receptor agonists. CBGV displayed enhanced potency in many of the functional outputs. However the most interesting result was a biased signaling that correlated with differential affinity, i.e. the overall results suggest that the binding mode of each ligand leads to specific receptor conformations underlying biased signaling outputs.
Conclusion: Results here reported and the recent elucidation of the three-dimensional structure of CB1 and CB2 receptors help understanding the mechanism of action that might be protective and the molecular drug-receptor interactions underlying biased signaling.”
https://pubmed.ncbi.nlm.nih.gov/32470563/
https://www.sciencedirect.com/science/article/abs/pii/S1043661820312482?via%3Dihub
 “Interest in CBG (cannabigerol) has been growing in the past few years, due to its anti-inflammatory properties and other therapeutic benefits.
“Interest in CBG (cannabigerol) has been growing in the past few years, due to its anti-inflammatory properties and other therapeutic benefits. 
 “Streptococcus mutans
“Streptococcus mutans “Preclinical studies have shown cannabidiol is protective in models of ischemic stroke. Based on results from our recent systematic review, we investigated the effects of two promising neuroprotective phytocannabinoids, cannabigerol (CBG) and cannabidivarin (CBDV), on cells of the blood-brain barrier (BBB), namely human brain microvascular endothelial cells (HBMECs), pericytes, and astrocytes.
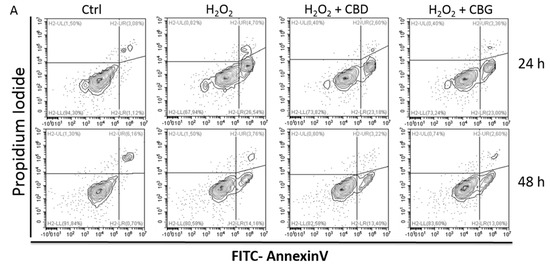
“Preclinical studies have shown cannabidiol is protective in models of ischemic stroke. Based on results from our recent systematic review, we investigated the effects of two promising neuroprotective phytocannabinoids, cannabigerol (CBG) and cannabidivarin (CBDV), on cells of the blood-brain barrier (BBB), namely human brain microvascular endothelial cells (HBMECs), pericytes, and astrocytes. “Glioblastoma is the most aggressive cancer among primary brain tumours. As with other cancers, the incidence of glioblastoma is increasing; despite modern therapies, the overall mean survival of patients post-diagnosis averages around 16 months, a figure that has not changed in many years. Cannabigerol (CBG) has only recently been reported to prevent the progression of certain carcinomas and has not yet been studied in glioblastoma. Here, we have compared the cytotoxic, apoptotic, and anti-invasive effects of the purified natural cannabinoid CBG together with CBD and THC on established differentiated glioblastoma tumour cells and glioblastoma stem cells. CBG and THC reduced the viability of both types of cells to a similar extent, whereas combining CBD with CBG was more efficient than with THC. CBD and CBG, both alone and in combination, induced caspase-dependent cell apoptosis, and there was no additive THC effect. Of note, CBG inhibited glioblastoma invasion in a similar manner to CBD and the chemotherapeutic temozolomide. We have demonstrated that THC has little added value in combined-cannabinoid glioblastoma treatment, suggesting that this psychotropic cannabinoid should be replaced with CBG in future clinical studies of glioblastoma therapy.”
“Glioblastoma is the most aggressive cancer among primary brain tumours. As with other cancers, the incidence of glioblastoma is increasing; despite modern therapies, the overall mean survival of patients post-diagnosis averages around 16 months, a figure that has not changed in many years. Cannabigerol (CBG) has only recently been reported to prevent the progression of certain carcinomas and has not yet been studied in glioblastoma. Here, we have compared the cytotoxic, apoptotic, and anti-invasive effects of the purified natural cannabinoid CBG together with CBD and THC on established differentiated glioblastoma tumour cells and glioblastoma stem cells. CBG and THC reduced the viability of both types of cells to a similar extent, whereas combining CBD with CBG was more efficient than with THC. CBD and CBG, both alone and in combination, induced caspase-dependent cell apoptosis, and there was no additive THC effect. Of note, CBG inhibited glioblastoma invasion in a similar manner to CBD and the chemotherapeutic temozolomide. We have demonstrated that THC has little added value in combined-cannabinoid glioblastoma treatment, suggesting that this psychotropic cannabinoid should be replaced with CBG in future clinical studies of glioblastoma therapy.” “Medical cannabis and individual cannabinoids, such as tetrahydrocannabinol (THC) and cannabidiol (CBD), are receiving growing attention in both the media and the scientific literature. The Cannabis plant, however, produces over 100 different cannabinoids, and cannabigerol (CBG) serves as the precursor molecule for the most abundant phytocannabinoids.
“Medical cannabis and individual cannabinoids, such as tetrahydrocannabinol (THC) and cannabidiol (CBD), are receiving growing attention in both the media and the scientific literature. The Cannabis plant, however, produces over 100 different cannabinoids, and cannabigerol (CBG) serves as the precursor molecule for the most abundant phytocannabinoids. “Fish oil (FO) and phytocannabinoids have received considerable attention for their intestinal anti-inflammatory effects.
“Fish oil (FO) and phytocannabinoids have received considerable attention for their intestinal anti-inflammatory effects. “Previous preclinical studies have demonstrated that cannabidiol (CBD) and cannabigerol (CBG), two non-psychotomimetic phytocannabinoids from Cannabis sativa, induce neuroprotective effects on toxic and neurodegenerative processes.
“Previous preclinical studies have demonstrated that cannabidiol (CBD) and cannabigerol (CBG), two non-psychotomimetic phytocannabinoids from Cannabis sativa, induce neuroprotective effects on toxic and neurodegenerative processes. “Background: Recent approved medicines whose active principles are Δ9Tetrahidrocannabinol (Δ9-THC) and/or cannabidiol (CBD) open novel perspectives for other phytocannabinoids also present in Cannabis sativa L. varieties. Furthermore, solid data on the potential benefits of acidic and varinic phytocannabinoids in a variety of diseases are already available. Mode of action of cannabigerol (CBG), cannabidiolic acid (CBDA), cannabigerolic acid (CBGA), cannabidivarin (CBDV) and cannabigerivarin (CBGV) is, to the very least, partial.
“Background: Recent approved medicines whose active principles are Δ9Tetrahidrocannabinol (Δ9-THC) and/or cannabidiol (CBD) open novel perspectives for other phytocannabinoids also present in Cannabis sativa L. varieties. Furthermore, solid data on the potential benefits of acidic and varinic phytocannabinoids in a variety of diseases are already available. Mode of action of cannabigerol (CBG), cannabidiolic acid (CBDA), cannabigerolic acid (CBGA), cannabidivarin (CBDV) and cannabigerivarin (CBGV) is, to the very least, partial.
 “Cannabidiol (CBD) and cannabigerol (CBG) are Cannabis sativa terpenophenols.
“Cannabidiol (CBD) and cannabigerol (CBG) are Cannabis sativa terpenophenols.